New Approach to Spatiotemporally and Globally Profile DNA–Protein Interactions Enabling Discovery of Low-Affinity Transcription Factors
DNA-protein interactions are essential components of biological systems, playing a crucial role in various biological processes. Precise dissection of DNA-protein interactions is essential for elucidating recognition basis, dynamics, and gene regulation mechanism. Despite of great progress being made in deciphering the codes of DNA-protein recognitions, global profiling of weak and dynamic DNA-protein interactions remains highly challenging.
Covalent capturing of DNA-protein interactions offers a promising and effective strategy to address such issues. However, current crosslinking strategies suffer from low efficiency, high non-specificity or insufficient interactome coverage, which affect sensitivity, accuracy and depth of the mass spectrometry (MS)-based proteomic profiling for the dynamic DNA-protein interaction networks.
In a study published in Nature Chemistry on April 27, a team of researchers led by Chen Xiao-hua and TAN Minjia from Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences developed a light-induced lysine (K) enabled crosslinking (LIKE-XL) strategy for spatiotemporal and global profiling of DNA-protein interaction, based on their recently developed PANAC photoclick reaction (Nat. Commun. 2020, Chem 2019) and expertise in quantitative proteomics. LIKE-XL enabled discovery of low-affinity transcription factors (TFs)-DNA interactions, determination of the binding sites for previously known TFs, and deciphering of the dynamics of TF subproteome in response to drug treatment in time-resolved manner.
As a proof-of-concept study, the researchers designed and synthesized the photo-reactive group-conjugated DNA probes, which enabled rapid and efficient capture of DNA-histone interactions. LIKE-XL, combined with MS-based quantitative proteomics, outperformed non-covalent pulldown regarding to identification number and MS intensities of captured interacting proteins (including TFs). Moreover, LIKE-XL showed much higher efficiency and sensitivity in profiling the specific DNA-protein interactions than non-specific crosslinking strategies. In addition to the expected low-affinity TF-DNA interactions, LIKE-XL enabled discovery of previously unknown TFs and their binding sites on the target DNA sequence; integration of crosslinking site data, structural information and molecular docking revealed the potential binding orientation of the TFs bound to the DNA probe. Furthermore, global profiling of DNA-TF interactions via LIKE-XL presents unexpected dynamics of TF sub-proteome in response to exogenous triggers (e.g. histone deacetylase inhibitor SAHA) in time-resolved manner.
By integrating the advantages of residue selectivity, spatiotemporal control, high efficiency and good biocompatibility, LIKE-XL strategy not only presents an effective and general method for constructing oligonucleotide-protein conjugates, but also provides a novel strategy to spatiotemporally and globally capture and identify the DNA-protein interactions with high sensitivity and accuracy. With these unique features, LIKE-XL enables discovery of low-affinity proteins via DNA sequences of interest and precise dissection of dynamic DNA binding activity profiles of proteins. In addition, the LIKE-XL offers a novel and complementary method to expand the DNA-centric profiling toolbox, to investigate DNA binding motifs previously unachieved for various proteins and to explore TFs-involved mechanism of action in drug perturbations. Moreover, as the feasibility and success demonstrated in global profiling DNA-TF interactions, the LIKE-XL is also a promising method to expand the repertoire of protein interactomes of DNA modifications. Therefore, this strategy will have an impact on a wide range of fields involving chemical biology, nucleic acid chemistry, proteomics, DNA and RNA epigenetics, bioconjugate chemistry.
DOI: https://doi.org/10.1038/s41557-023-01196-z
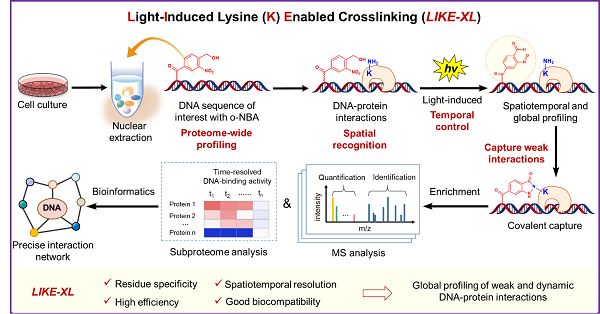
The LIKE-XL strategy for the spatiotemporal and global profiling of DNA–protein interactions (Image by Hao Hu)




