Researchers Reveal Comprehensive Proteogenomic Landscape of Human Small Cell Lung Cancer
Small cell lung cancer (SCLC) is the most malignant and deadliest subtype of lung cancer, which kills over 200,000 people worldwide each year. Despite progress has been achieved at multiple fronts in the ‘war’ on this cancer, clinical outcomes for SCLC patients have shown only modest improvement. Gaining comprehensive understanding of its tumor biology is imperative.
In a study published in Cell on Jan 4, a team of Chinese researchers led by ZHOU Hu from Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences, in collaboration with groups led by ZHANG Peng from Shanghai Pulmonary Hospital of Tongji University, and JI Hongbin and GAO Daming from Shanghai Institute of Biochemistry and Cell Biology of Chinese Academy of Sciences, reported comprehensive proteogenomic landscape of SCLC tumors and provides new insights of tumor biology of this recalcitrant cancer.
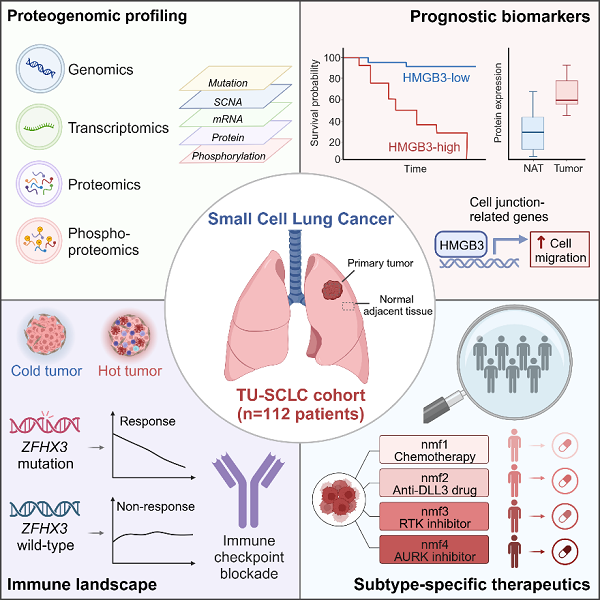
In this study, researchers performed an integrated genomic, transcriptomic, proteomic, and phosphoproteomic analysis on 112 tumors and paired adjacent lung tissues resected from Chinese SCLC patients. Combining the multi-omics data with clinical information, researchers described a comprehensive molecular landscape of SCLC.
Leveraging integrated multi-omics analysis, researchers explored the functional consequences of genomic aberrations. They found that FAT1 mutation led to loss of function and may promote an epithelial mesenchymal transition (EMT) phenotype in SCLC. They also discovered that RB1 copy number loss impact SCLC’s proteomes and phosphoproteomes via trans-effects.
Researchers identified two prognostic protein biomarkers, HMGB3 and CASP10 by proteomic-based differential analysis. High expression of HMGB3 was linked to worse survival, while CASP10 was associated with better prognosis. Further biological experiments indicated that HMGB3 also functions as an oncogenic factor in SCLC via transcriptional regulation of cell junction-related genes.
Notably, the researchers observed ZFHX3 mutation was enriched in “immune-hot’’ tumors and showed a promising correlation to major pathological responses after experimental neoadjuvant chemo-immunotherapies. Thus, they indicated that ZFHX3 mutation could be a potential predictive biomarker for SCLC patients receiving immunotherapy.
In addition, researchers refined all the tumors into four multi-omic subtypes with distinct molecular features. Chemotherapy, DLL3-targeting agents, receptor tyrosine kinase (RTK) inhibitors, and aurora kinases (AURK) inhibitors are nominated as potential individualized treatments for each of the multi-omic subtypes. This is supported by in vivo drug response test based on SCLC patient-derived xenograft (PDX) or SCLC cell line-derived xenograft (CDX) models.
This study extends the understanding of SCLC cancer biology and generates subtypes that may guide precision therapeutics. The underlying dataset also furnish a valuable resource for basic and clinical researchers and will contribute to more effective SCLC clinical treatment.
DOI: https://doi.org/10.1016/j.cell.2023.12.004
proteogenomic landscape of small cell lung cancer (Image by SIMM)
Contact:
DIAO Wentong
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: diaowentong@simm.ac.cn




