Spatiotemporal and Direct Capturing Global Substrates of Post-Translational Modifying Enzymes in Living Cells
Protein-modifying enzymes regulate the dynamics of myriad post-translational modification (PTM) substrates. Precise characterization of enzyme-substrate associations is essential for understanding the molecular basis of cellular function and phenotype. Since protein-modifying enzymes usually control the dynamics for numerous PTM substrates, global profiling of substrates could dissect the complicated and elusive enzyme-substrate interactions, and further illuminate the landscape of complex regulatory networks. Despite recent advancements, the global profiling of direct substrates for PTM-enzymes in living cells still faces several challenges, such as loss of weak and transient interactions, inability to precisely distinguish the direct and indirect PTM-substrates/sites. In principle, covalently capturing enzyme-substrate interactions offers a promising and effective strategy to address such issues. Although site-specifically incorporating unnatural amino acid crosslinkers into proteins is widely used to capture protein-protein complexes, the potential and capability of PTM-mechanism-guided direct capturing global substrates presents an exciting prospect but yet largely unexplored.
In a study published in Nature Communications on February 17, researchers led by Xiao-Hua Chen and Minjia Tan from the Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences, introduced a novel strategy to directly capture substrates of PTM-enzymes via PTM-acceptor residue crosslinking in living cells. This method was based on their recently breakthroughs, which developed light-induced primary amines and o-nitrobenzyl alcohols cyclization (PANAC) chemistry (Nat. Commun. 2020, Chem 2019, Nat. Chem. 2023, 15, 803). The utilization of this strategy allows for spatiotemporal and global profiling of direct substrates of PTM-enzymes and facilitating the validation of PTM-sites in a straightforward manner.
By integrating enzymatic PTM-mechanisms and genetically encoding residue-selective unnatural amino acid o-NBAK into PTM-enzymes, the strategy successfully achieved the direct capturing and global profiling of the substrates for both bacterial and mammalian lysine acylation enzymes, including bacterial lysine acylases PatZ, YiaC, LplA, TmcA, and YjaB, as well as mammalian acyltransferases GCN5 and Tip60. Notably, this approach broadened the substrate profiles of these investigated enzymes, leading to discovery of functionally important new substrates and acylation sites. For example, in this study, the researchers identified that acetylation on K131 of AcrA regulates the assembly of AcrA-AcrB-TolC multidrug efflux pump, suggesting its potential regulatory role in antibiotic resistance.
With high residue-selectivity, spatiotemporal controllability, covalent capture of the PANAC photoclick chemistry introduced into the native PTM-enzymes in living cells, the direct substrate-capturing strategy enables decoding substrates with heightened sensitivity and accuracy, as well as straightforward validation of acylation sites. The exploration of global substrate profiles advanced the understanding of PTM-enzymes mechanisms, potential substrates, and protein functions. The discovery and functional validation of new substrates provided fresh insights into the molecular mechanisms underlying related diseases, along with novel avenues for modulating the dynamic interactions of proteins. More importantly, harnessing the recent advances on other kinds of residue-selective crosslinking chemistry, the concept of direct capturing substrates of PTM-enzymes developed in current research could inspire and lead to further development of new capturing strategies for other types of PTM-enzymes. This strategy would expand the repertoire of enzyme-centric methods for investigating more PTMs and the landscape of enzyme-substrate interactive proteomics.
DOI: https://doi.org/10.1038/s41467-024-45765-3
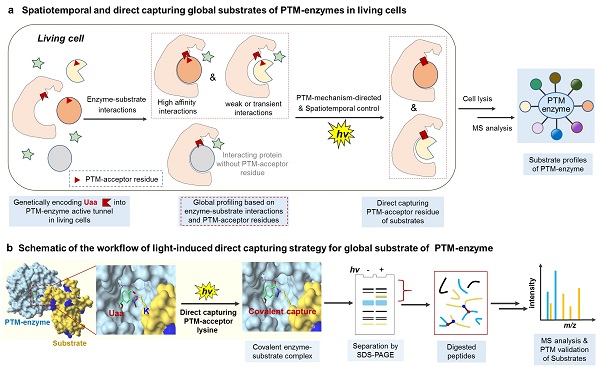
Spatiotemporal and direct capturing global substrates of PTM-enzyme via PTM-acceptor residue of substrates. (a) Genetically encoding residue-selective unnatural amino acid (Uaa) o-NBAK in living cells, offering global substrate profiles of PTM-enzymes from MS analysis. (b) Schematic of the workflow of light-induced direct capturing strategy for global substrate of PTM-enzyme. (Image by Hao Hu)
Media Contact:
JIANG Qingling
Shanghai Institute of Materia Medica
qljiang@stimes.cn




