Scientists Establish integrative Proteogenomic and Pharmacological Landscape of Acute Myeloid Leukaemia in Chinese Population
Acute myeloid leukaemia (AML) is mainly characterized by an increase in myeloid cells in bone marrow and a decrease in mature cells, accounting for 28% of leukaemia cases, with a five-year survival rate of only 30.5%. AML has the lowest median mutation frequency among hematologic malignancies, reflecting the potential of molecular heterogeneity at transcriptional and translational level. Quantitative proteomics analyses uncovered the heterogeneity of Western AML patients at the protein and phosphorylation modification levels. However, drug sensitivity studies integrated with a more comprehensive multi-omics characterization are still lacking, which could promote a deeper understanding of molecular characteristics of AML and their relationship to drug responses. Moreover, the proteomics of Asian populations is still poorly understood.
In a study published in Science Bulletin, a research team led by Profs. LI Jia, ZHOU Yubo, TAN Minjia from the Shanghai Institute of Materia Medica of the Chinese Academy of Sciences, along with Prof. YANG Jianmin from Naval Medical University, presented a comprehensive genomic, proteomic and phosphoproteomic analysis of 101 samples from Chinese AML patients and a systemic in vitro drug sensitivity analysis of 77 drugs. Proteome-based unsupervised clustering revealed three subtypes with different molecular characteristics and clinical outcomes. Further integrative analysis of drug sensitivity combined with proteomic/phosphoproteomic data uncovered potential drug combinations.
The study employed consensus clustering to identify AML subtypes. The results showed that three subtypes were more considerable than two or four subtypes. Subtype S-I was characterized by higher tumour proliferation, alterations in carbohydrate and nucleic acid metabolism, such as proteasome, actin cytoskeleton regulation, and leukocyte transendothelial migration. Phagosomes, lysosomes, and ferroptosis were enriched in subtype S-II group. In contrast, subtype S-III was distinguished by several metabolic pathways and RNA processes, such as fatty acid metabolism, TCA cycle, and oxidation phosphorylation. The study also found that the Measurable Residual Disease (MRD) after treatment was higher in subtype S-I, reflecting lower clearing efficiency.
The study showed that allogenic haematopoietic stem cell transplantation (Allo-SCT) did not prolong survival of patients with subtype S-I. However, patients with subtypes S-II&III significantly benefited from Allo-SCT. These results indicated that Allo-SCT might be a potential treatment strategy for subtypes S-II&III. In addition, patients with subtype S-I had high expression of XPO1 protein, and in vitro drug sensitivity results showed that they were more sensitive to the XPO1 inhibitor KPT-185. These findings could help guide recommendations for Allo-SCT and alternative therapies.
To further understand the correlation between drug sensitivity and molecular characteristics, researchers profiled primary tumour cells from 56 patient samples against a panel of 77 inhibitors involving clinical chemotherapy drugs, kinase inhibitors and epigenetic inhibitors .
The study showed that higher expression of ALDH3A2 was positively correlated with the cytarabine sensitivity value. Researchers treated 10 AML cell lines with a combination of ALDH3A2 inhibitor disulfiram and cytarabine, finding that disulfiram could sensitize therapeutic efficacy of cytarabine in most of the cell lines. In addition, the multi-omics data helped investigate the underlying molecular characteristics of Acalisib (PI3K inhibitor) resistance. In the Spearman correlation analysis of drug sensitivity and the abundance of a single phosphosite, the study found that high kinase activation of PDK was related to low Acalisib sensitivity. Researchers also found that the PDK inhibitor GSK2334470 could improve the anticancer effects of Acalisib and another PI3K inhibitor, GDC0032.
In conclusion, the multi-omic integrative analysis revealed valuable insight for linking molecular characterizations with clinical outcomes. The study also provided an additional molecular layer beyond genetic and proteomic characterization of AML, offering potential diagnosis and therapeutic strategies. The biomarkers discovered based on this multi-omics landscape could help provide clues and suggestions for subsequent diagnosis and treatment of AML.
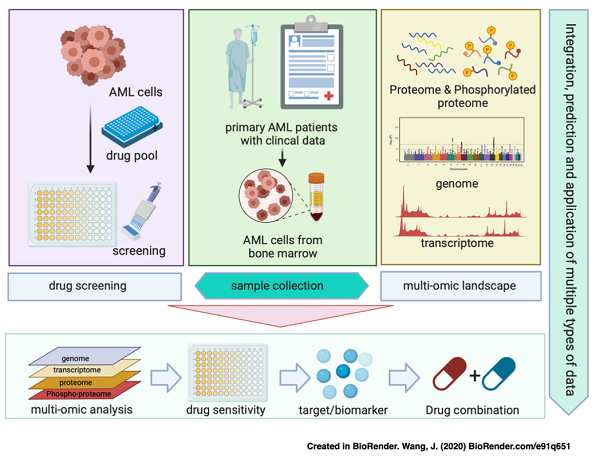
Graphic abstract: the establishment process of AML multi-omics database and drug sensitivity analysis platform (Image by WANG Hanlin)
Contact:
JIANG Qingling
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: qljiang@stimes.cn




