Scientists Discover Lysine-Targeting Covalent Inhibitors via DNA-Encoded Chemical Library Selection Platform Guided by Proteome-Wide Data
Covalent drugs function by forming covalent bonds with specific amino acid residues, enabling sustained modulation of target proteins. This therapeutic modality has become a critical focus in modern drug discovery. Compared with cysteine-targeting strategies, lysine serves as an alternative covalent binding site that circumvents the limitation of cysteine scarcity in ligand-binding pockets and broadens the landscape of druggable targets. In recent years, structure-based drug design has facilitated the development of lysine-targeting covalent inhibitors. However, these approaches frequently depend on known ligand scaffolds, limiting their applicability to novel or traditionally undruggable targets.
The covalent DNA-encoded chemical library (CoDEL) technology is rapidly emerging as a key platform for covalent drug discovery. The group led by LU Xiaojie from the Shanghai Institute of Materia Medica of the Chinese Academy of Sciences, in collaboration with others, has applied this technology to discover novel cysteine-targeting covalent inhibitors in proteins such as BTK, JAK3, PIN1, and SARS-CoV-2 non-structural proteins. Furthermore, LU’s group developed an integrated Activity-Based Protein Profiling (ABPP)-CoDEL strategy to identify tyrosine-targeting covalent inhibitors. However, a systematic CoDEL selection platform specifically designed for lysine-targeting inhibitors has yet to be established. Additionally, the widespread distribution of lysine across the human proteome makes random target selection inefficient for screening.
In a study published in Angewandte Chemie International Edition on April 13, a research team led by LU, in collaboration with ZHOU Lu from Fudan University and SUN Yi from the Cancer Institute of the Second Affiliated Hospital of Zhejiang University School of Medicine and Zhejiang University Institute of Translational Medicine, integrated activity-based protein profiling (ABPP) data with CoDEL technology, and identified structurally novel lysine-targeting covalent inhibitors with diverse mechanisms of action.
By integrating compound-based and warhead-based ABPP datasets, the research team constructed a protein dataset enriched with lysine residues exhibiting both high reactivity and ligandability, thereby facilitating rational target selection for screening. Subsequently, 8 lysine-targeting covalent warheads with distinct reaction mechanisms were incorporated to synthesize CoDELs comprising 10.7 million compounds. Covalent selection identified lysine-targeting covalent inhibitors against phosphoglycerate mutase 1 (PGAM1), bromodomain (BRD) family proteins, and ubiquitin-conjugating enzyme E2 N (UBE2N). Among them, Compound 1 functioned as a photo-covalent probe for the active site of PGAM1, while Compound 4 formed a reversible covalent bond with a previously unexplored site within the bromodomain of BRD family proteins. Notably, Compound 9 irreversibly bound to UBE2N, induced conformational changes in the UBE2N/UBE2V2 complex, disrupted polyubiquitin chain formation, and impaired its downstream functional activity. This novel mechanism offered a new strategy for regulating the ubiquitination pathway.
This study established an efficient selection platform for lysine-targeting covalent inhibitors by integrating proteomic data with CoDEL technology. This strategy not only expanded the applicability of covalent drugs in target selection but also provided new insights and technical support for the rational design of covalent inhibitors.
Link: https://doi.org/10.1002/anie.202505581
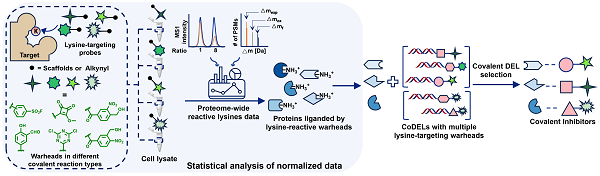
The proposed proteome-wide data guide the lysine-targeting CoDEL selection strategy. (Image by WU Xinyuan)
Contact:
JIANG Qingling
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: qljiang@stimes.cn




