Scientists Discover Residue-Selective Inhibitors via Covalent DNA-Encoded Chemical Library Selection with Diverse Warheads
Covalent drugs specifically target key cancer sites by forming covalent bonds with residues on the targets, making them important strategies for precision cancer therapy. With the advancement of chemical biology and mass spectrometry technologies, covalent targeting strategies have expanded to include nine different amino acid residues. In addition to cysteine, lysine, serine, and other residues have emerged as new targets due to their high abundance. Moreover, covalent modifications of weakly reactive residues such as aspartic acid and histidine have also been reported. However, efficiently identifying covalent inhibitors targeting different amino acids remains a key challenge in covalent drug discovery.
The covalent DNA-encoded chemical library (CoDEL) technology enables rapid synthesis of billions of covalent compounds, generating a compound library much larger than commercially available libraries. This makes CoDEL an ideal platform for integrating electrophilic warheads and performing residue-specific targeting selection.
The group led by LU Xiaojie from the Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences, in collaboration with others, has applied CoDEL to discover cysteine-targeting covalent inhibitors. Moreover, the application range of this technology was expanded to include covalent inhibitors targeting tyrosine and lysine through an Activity-Based Protein Profiling (ABPP)-CoDEL combined strategy. Further exploration of CoDEL's capabilities in screening covalent inhibitors across a broader range of amino acid residues may provide an efficient and widely applicable strategy for developing residue-selective covalent drugs.
In a study published in Journal of the American Chemical Society on April 28, a research team led by LU, in collaboration with AI Jing and TAN Minjia from SIMM, established a CoDEL selection platform that integrated multiple electrophilic warheads targeting different amino acid residues. Using Fibroblast Growth Factor Receptor 2 (FGFR2) as a target, active covalent inhibitors targeting cysteine, lysine, arginine, or glutamic acid were identified. Among these, arginine-targeting covalent inhibitors exhibited excellent subtype selectivity and resistance to drug-resistant mutations. This platform broadened the scope of rational design for covalent inhibitors and provided a method to mitigate the impact of target mutations while meeting specificity requirements, enabling more precise therapeutic interventions.
FGFR kinase family proteins are critical targets for cancer therapy. Selective inhibitors of FGFR2 have shown good safety in clinical applications but are challenged by acquired resistance due to residue mutations. In this study, a reactivity-based evaluation was conducted using 17 covalent warheads targeting 9 amino acid residues in FGFR2, resulting in the construction of a CoDEL library containing 24.8 million compounds. Covalent FGFR2 inhibitors targeting cysteine, lysine, arginine, or glutamic acid were identified. The cysteine-targeting covalent inhibitor CF3 exhibited potent inhibition of FGFR2. The lysine-targeting covalent inhibitor KF7 revealed a previously uncharacterized reactive site, overcoming limitations of targeting conventional sites. The arginine-targeting covalent inhibitor RF3 was found to be a highly selective FGFR2/3 inhibitor, demonstrating significant inhibition of the FGFR2 V564F gatekeeper mutation. Additionally, the glutamic acid-targeting covalent inhibitor AF1 confirmed glutamic acid as a viable covalent drug binding site, laying the foundation for future development of more effective covalent inhibitors.
This work demonstrated the feasibility of discovering residue-specific covalent inhibitors using a multi-warhead CoDEL selection platform. This strategy not only expanded the target space for covalent drugs but also enhanced the application potential of CoDEL technology in precision therapy, providing new insights and effective approaches for drug development.
DOI: 10.1021/jacs.5c01712
Link: https://doi.org/10.1021/jacs.5c01712
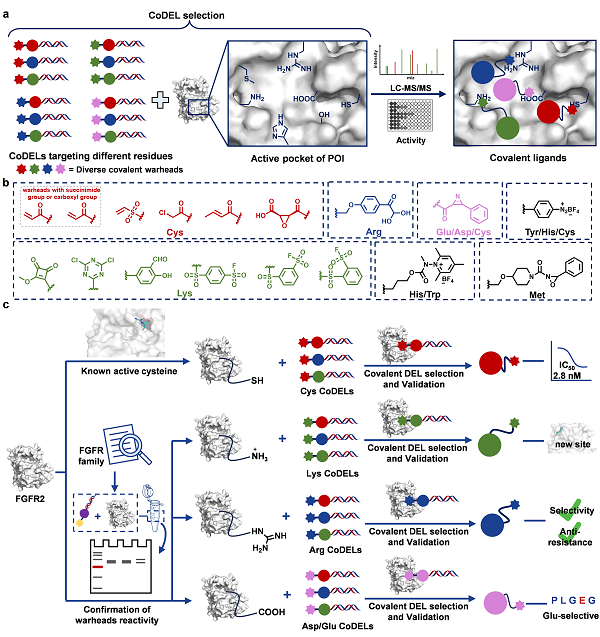
The proposed strategy of residue-selective covalent inhibitors discovery through CoDELs. (Image by WU Xinyuan)
Contact:
JIANG Qingling
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: qljiang@stimes.cn




