NMRExtractor: Leveraging Large Language Models to Construct an Experimental NMR Database from Open-source Scientific Publications
Understanding and predicting molecular structures from spectroscopic data is fundamental to both chemistry and drug discovery. Nuclear Magnetic Resonance (NMR) spectroscopy remains one of the most informative techniques for structure elucidation. However, the lack of structured, high-quality NMR datasets hinders its integration with AI tools.
In a study published in Chemical Science on May 28, a research team led by Prof. ZHENG Mingyue from the Shanghai Institute of Materia Medica of the Chinese Academy of Sciences introduced NMRExtractor, a large language model (LLM)-based method for automated NMR data extraction from chemical literature.
In this work, the researchers developed NMRExtractor by fine-tuning Mistral-7B-instruct LLM on a curated dataset of human-annotated NMR examples. The model was optimized to interpret complex textual descriptions of NMR results in open-access PubMed articles and output structured data, including 1H and 13C chemical shifts, experimental conditions, and associated molecular metadata. With an extraction accuracy exceeding 96% on standard inputs, NMRExtractor substantially outperforms existing rule-based or regular-expression approaches.
The researchers further constructed NMRBank, a comprehensive database containing over 225,000 high-confidence NMR records extracted from 5.7 million articles. Each record includes SMILES strings, IUPAC names, solvent and field conditions, chemical shift values, and confidence scores. Compared to legacy datasets such as NMRShiftDB2, NMRBank significantly broadens coverage in terms of molecular weight, topological polar surface area (TPSA), and lipophilicity (logP), thereby enriching the chemical diversity available for downstream AI model training and NMR prediction tasks.
NMRExtractor’s architecture includes a confidence estimation module that enables users to select high-reliability records (confidence >0.8) for constructing machine-learning-ready datasets. Notably, its performance remains robust even when facing irregular or nonstandard NMR reporting formats, which are frequently encountered in published literature.
The method’s ability to generalize across diverse textual styles and domains highlights the potential of LLMs to transform scientific knowledge extraction at scale. By bridging the gap between unstructured literature and structured NMR repositories, NMRExtractor and NMRBank offer valuable tools for the chemistry, pharmaceutical, and data science communities.
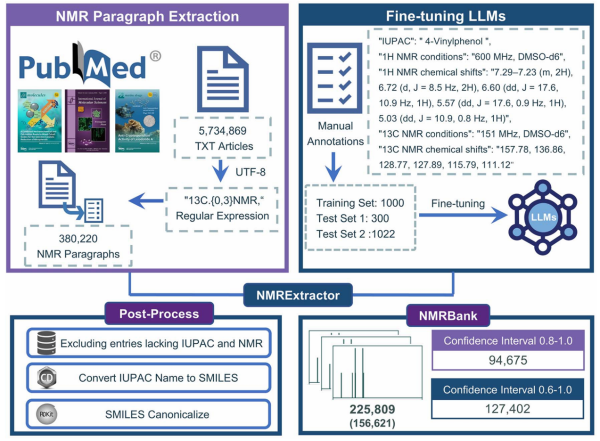
Schematic diagram of the NMRExtractor extraction process and the construction of the NMRBank dataset. (Image by ZHENG’s lab)
Contact:
JIANG Qingling
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: qljiang@stimes.cn




