Researchers Reveal a Pressure Sensing and Modulating Mechanism Underpinning Genome Packaging, Retention and Ejection in Herpesviruses
Human cytomegalovirus (HCMV), the prototypical member of the β-herpesvirinae within the Herpesviridae, is found in about 90% of the population worldwide. HCMV infection can lead to severe and sometimes life-threatening diseases in immunocompromised individuals. HCMV is also the leading viral cause of congenital infections that can lead to birth defects. Herpesviruses package the genome via a “head-full” mechanism. The portal, which is both the entering and exiting channel of the viral DNA, has been suggested to serve as the “head-full” pressure sensor. The genome size (235 kb) of HCMV is the largest among the known human herpesviruses albeit the capsid size of HCMV is similar to those of other herpesviruses. Given that the genome sizes of herpesviruses vary a lot, it is still unknown how the headful of genome package among the different herpesviruses is sensed by the portals.
In a study published online in Nature Communications on July 27th, researchers of Dr. YU Xuekui’s group at the Shanghai Institute of Materia Medica of the Chinese Academy of Sciences resolved the in situ structures of the portal and the capsid vertex-specific components (CVSC) of HCMV, which provides structural basis underpinning the genome packaging, retention, and ejection of herpesviruses.
Through the method of improved viral growth, the researchers obtained high quality viral sample and determined the in situ cryo-electron microscopy (CryoEM) structures of the portal and the CVSC of HCMV. The portal of HCMV has three symmetry mismatches: the 5-fold symmetric (C5) 10-helix anchor, the 6-fold symmetric (C6) turret and the 12-fold symmetric (C12) main body. The 10-helix anchor—uncommon among known portals—contacts the portal-encircling DNA, which was presumed to squeeze the portal as the genome packaging proceeds. Researchers surmise that the 10-helix anchor dampens this action to delay the portal reaching a “head-full” packaging state, thus facilitating the large genome to be packaged. The 6-fold symmetric turret, latched via a coiled coil to a helix from a major capsid protein, supports the portal to retain the packaged genome. In addition, researchers found the portal vertex undergoing conformational changes simultaneously to secure the genome during viral cell trafficking and to facilitate genome ejection when nucleocapsid reaches the nucleus.
Also, researchers determined the stoichiometric number of the CVSC bound on the penton vertices of HCMV, and found that the peripenton CVSC occupancy in HCMV is significantly lower than that in HSV-1, KSHV, and EBV; albeit, HCMV contains the largest DNA genome.
This research provided insight into the conservation and adaption of HCMV portals and outline how the capsid inner pressure is sensed and modulated among herpesviruses to accomplish the tasks of genome packaging, retention, and ejection.
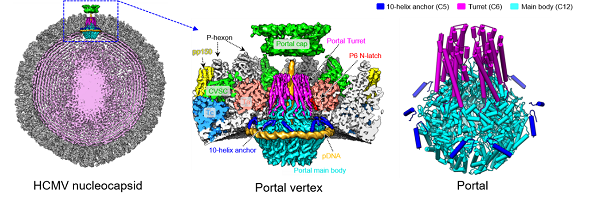
Figure. Left, cryoEM structure of the HCMV nucleocapsid. Middle, cryoEM structure of portal vertex of HCMV. Right, atomic model of the symmetry-mismatched portal of HCMV. (Image by Li Zhihai of YU Xuekui’s group)
DOI: https://doi.org/10.1038/s41467-021-24820-3
Link: https://www.nature.com/articles/s41467-021-24820-3
Contact:
DIAO Wentong
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: diaowentong@simm.ac.cn




