Proteomics of Adjacent-to-Tumor Samples Uncovers Clinical Biological Event in Hepatocellular Carcinoma
Primary liver cancer is the fourth most common and second highest mortality malignant tumor in China. Hepatocellular carcinoma (HCC) comprises 75%–85% of liver cancer cases and is of high heterogeneity. Most oncology studies focus on tumor tissues, and few in-depth studies on normal adjacent tissues (NATs) are carried out. NATs, also known as non-tumor liver tissues, are usually considered as control samples in tumor-related studies. The comprehensive analysis of transcriptome and genome-derived haplotype-specific somatic copy number alterations suggest that NAT is a unique intermediate state between healthy tissue and tumor and may accumulate oncogenic events. However, the comprehensive analysis of NATs at the proteomic level remains less studied.
In a study published in National Science Review (NSR) in June, ZHOU Hu’s team from Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences, in collaboration with FAN Jia and GAO Qiang’s team from Zhongshan Hospital, Fudan University, performed a systematic analysis of the molecular heterogeneity in HCC NATs at the proteomic level and deciphered their proteomic differences compared to healthy livers.
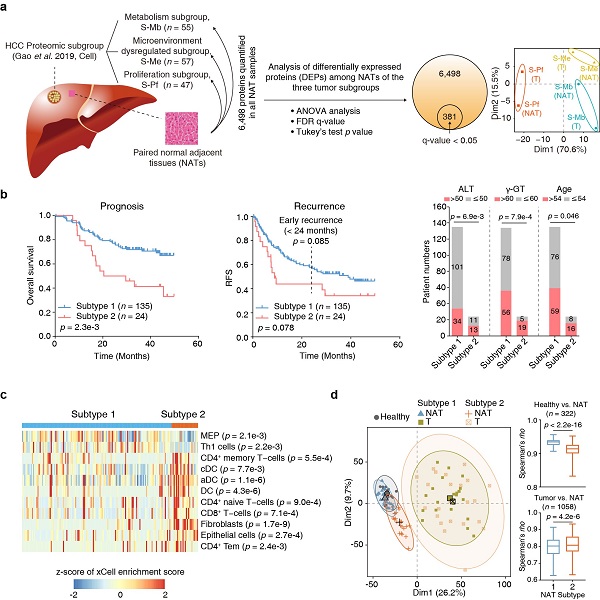
The researchers previously divided HCC into three tumor subgroups, namely metabolism subgroup, microenvironment dysregulated subgroup and proliferation subgroup (Cell, 2019), and proteomic analysis of the paired NATs of the three subgroups found that they had the molecular characteristics of the corresponding cancer tissues (Figure a). Unsupervised clustering divided the HCC NATs into two subtypes (Subtype 1, n = 135; Subtype 2, n = 24). Patients of Subtype 2 NATs accounted for about 15% of the total number of patients, with older age, and were associated with poor prognosis and high-risk recurrence. Association analysis of the clinical information with NAT subtypes suggested higher levels of aminoleucine transferase (ALT) and γ-glutamyl transferase in NAT subtype 2. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis suggested that the upregulated proteins in subtype 1 were enriched in pathways including ECM-receptor interaction, antigen processing and presentation, focal adhesion and PI3K-Akt signaling pathways. Downregulated proteins were enriched in various metabolic pathways. The NAT subtyping criteria above was further verified in another high-quality proteomic dataset of HCC NATs (Nature, 2019) (Figure b).
This study also uncovered clear differences in immune features between these two subtypes. The Subtype 2 NATs had significantly higher proportions of dendritic cells (DC), CD8+ T cells and fibroblasts, which were further validated by multiplexed immunofluorescence using tissue microarray (Figure c).
Moreover, the proteomic analysis using healthy liver tissues and the two NAT subtypes and their paired tumors by data-independent acquisition mass spectrometry (DIA-MS) further uncovered early molecular alterations associated with the progression from healthy livers to NATs. Hierarchical clustering analysis (HCA) and principal component analysis (PCA) suggested that Subtype 1 NATs were more similar to healthy liver tissues, whereas Subtype 2 NATs were much closer to the tumors. A total of 183 differently expressed proteins (DEPs) were identified between Subtype 1 NATs and healthy liver tissues, including 112 upregulated and 71 downregulated proteins. Among the upregulated proteins, 13 DEPs were annotated as secreted proteins, which may be candidates for early diagnosis biomarkers for fibrosis or HCC. The upregulated DEPs may also contain factors that have functions in changing tumor microenvironment and promoting tumor recurrence (Figure d).
The molecular heterogeneity of normal adjacent tissues in hepatocellular carcinoma (image by Zhu Hongwen)
DOI: https://doi.org/10.1093/nsr/nwad167/7189878
Contact:
DIAO Wentong
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: diaowentong@simm.ac.cn




