Researchers Reveal Dynamic Regulation of Ubiquitin Ligase E6AP Activity
E6AP, the founding member of the HECT-type ubiquitin ligase family, was initially discovered for its interaction with the oncogenic protein E6 of human papillomavirus (HPV). E6 from high-risk HPV hijacks the ubiquitin ligase activity of E6AP, leading to abnormal ubiquitination and degradation of the tumor suppressor p53. This process is associated with the development of various HPV-positive cancers, including over 90% of cervical cancers. HPV E6 not only alters the substrates of E6AP but also enhances its activity through an unknown mechanism. Additionally, deficiencies or enhancements in E6AP activity are associated with Angelman syndrome and autism spectrum disorders, respectively. Therefore, studying the regulatory mechanism of E6AP activity is of significant importance.
In a study published in Nature Communications on April 26, a team of researchers led by YU Xuekui and LUO Cheng from Shanghai Institute of Materia Medica (SIMM) of the Chinese Academy of Sciences, resolved the structures of E6AP and E6-bound E6AP using cryo-electron microscopy (cryo-EM). Structural comparison between the E6AP alone and the E6AP/E6 complex, complemented by molecular dynamics simulations and biochemical analysis, revealed the functional mechanism of E6AP.
In this study, researchers found the E6AP/E6 complex exhibited as a homodimer of heterodimer (protomer). Each protomer consists of one E6AP and one E6 molecule. The oncogenic protein E6 binds to a surface pocket, formed by the conserved α1-helix and a loop-helix-loop element of E6AP. The dimerization of E6AP/E6 protomers is primarily mediated by hydrophobic interactions between the carboxy termini of the two conserved α1-helices.
Additionally, the researchers obtained five different conformations of E6AP/E6 complex, and found that the protomers swing around the crossed region of the two α1-helices. The dynamic characters of the E6AP-E6 complex favor the proximity of E6AP’s C-lobe to the ubiquitin-conjugating enzyme E2 for accepting ubiquitin or to substrates for donating ubiquitin. Functional analysis further confirmed that the dynamics and dimerization of the E6AP/E6 protomer are required for E6AP effectively transferring ubiquitin to its substrates. Furthermore, the researchers found the α1-helix in E6AP alone exists as a short helix, and the counterpart in the E6AP/E6 complex became a longer helix. Molecular dynamics simulations showed that the extension of the α1-helix is stabilized by E6.
Based on these findings, this study proposed a dynamic regulatory mechanism of E6AP activity by E6: E6 binding stabilizes the elongation of E6AP α1-helix, thereby promoting the dimerization of E6AP and transforming the inactive E6AP into the active dimer. Actually, some E6AP mutations associated with Angelman syndrome and autism spectrum disorders are located in the extended α1-helix. The effects of these mutations are correlated with the stability or interactability of the extended α1 helix, highlighting the importance of the α1-helix in mediating dimerization interactions. The results of this study provide crucial structural information for understanding the physiological and pathological mechanisms of E6AP and contribute to the development of therapeutic interventions for diseases associated with E6AP dysfunction.
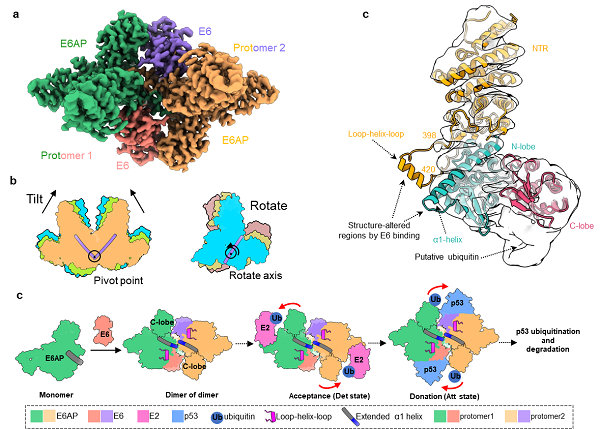
a. Overall structure of the E6AP/E6 complex (represented by the Att1 state, top view).
b. Schematic comparison of the five conformations of the E6AP/E6 complex.
c. Comparison between the atomic model of E6AP in the E6AP/E6 complex and the electron density map of monomeric E6AP.
d. Schematic diagram of the mode by which HPV E6 hijacks the enzymatic activity of E6AP.
DOI: https://doi.org/10.1038/s41467-024-47586-w
Media Contact:
JIANG Qingling
Shanghai Institute of Materia Medica, Chinese Academy of Sciences
E-mail: qljiang@stimes.cn




