Covalent DNA-Encoded Library Workflow Drives Discovery of SARS-CoV-2 Non-structural Protein Inhibitors
Covalent inhibitors are small molecules that form covalent bonds with nucleophilic residues, such as cysteines, in target proteins. Their unique interactions offer advantages such as enhanced potency, prolonged action, and high selectivity, particularly when targeting unique binding pockets or reactive residues. Covalent inhibitors have achieved significant progress in drug development, with applications spanning various therapeutic targets. High-throughput screening of covalent inhibitors has traditionally relied on commercial covalent fragment libraries and activity-based protein profiling (ABPP).
In a study published in Journal of the American Chemical Society (JACS) on November 22, 2024, a research team led by Prof. LU Xiaojie and Prof. XU Yechun from the Shanghai Institute of Materia Medica (SIMM) of Chinese Academy of Sciences, introduces a novel covalent DNA-encoded library (DEL) workflow. This method enables covalent screening against cysteine-rich non-structural proteins of SARS-CoV-2, leading to identification of covalent hit compounds with novel scaffolds and allosteric binding modes.
The researchers focused on SARS-CoV-2 non-structural proteins. Structural analyses identified three key targets: Nsp3 (PLpro), Nsp5 (3CLpro), and the cysteine-rich Nsp7/8/12 complex. The covalent DEL workflow was specifically designed by introducing a covalent linker as the final building block during library synthesis, enabling selective and rational design to target deeply buried reactive residues.
Using wild-type and catalytically inactive (Cys-mutant) protein variants, the team conducted comparative screenings to identify covalent inhibitors targeting catalytic residues. For the first time, researchers discovered compounds binding at an allosteric site outside the ubiquitin pocket of PLpro. Additionally, by correlating IC50 values with standardized enrichment, they predicted activity across diverse scaffolds. For 3CLpro, precise docking was achieved by introducing conformational constraints to DNA-linked compounds exposed to solvent regions, and this was further validated by protein-ligand crystal structures. Covalent analysis of Nsp7/8/12 was performed via gel electrophoresis and quantum mechanics/molecular mechanics (QM/MM) simulations, revealing potential binding sites.
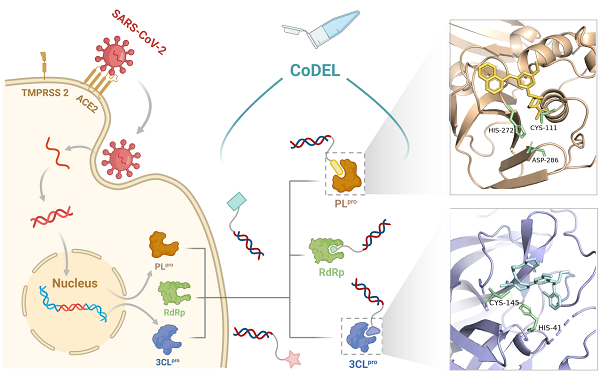
This study integrated DEL technology with covalent strategies, achieving efficient screening of SARS-CoV-2 non-structural proteins through rational design of covalent DELs. (Image by WANG Xudong)




