Study Develops Human-Derived AMBER Editor for Efficient C-to-U RNA Editing in vivo
In a study published in National Academy of Sciences of the United States of America (PNAS), a research team led by HAO Pei from the Shanghai Institute of Materia Medica, Chinese Academy of Sciences, and LI Xuan from the Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, reported the development of a novel RNA single-base (C-to-U) editing tool, termed ADAR2-Mimic C-to-U Base Editor for RNA (AMBER). They demonstrated that AMBER emulates the mode of natural adenosine deaminase acting on RNA (ADAR) and achieves precise C-to-U editing of target RNAs in cultured cells and mouse liver. The research showed that AMBER exhibits high editing efficiency, low immunogenicity and minimal off-target effects at the transcriptome level.
Existing C-to-U RNA editing tools face several challenges, including the risk of concurrent DNA mutations caused by natural APOBEC family, and the large-size anchoring domains associated with immunogenicity due to microbial origin of Cas13 protein. These issues restrict their potential for clinical translation.
To address these challenges, the research team chose human ADAR2 as a starting point. They introduced 17 key amino acid substitutions previously reported in the RESCUE-S editing system into the catalytic domain of ADAR2. Instead of fusing to an external RNA-targeting module, AMBER uses the native double-stranded RNA binding domain (dsRBD) of ADAR2 for target recognition. This design generates a fully human-derived C-to-U RNA editing system that does not rely on bacterial Cas proteins and is expected to have lower immunogenicity.
The researchers first tested AMBER with a reporter based on a mutant enhanced green fluorescent protein (eGFP). With the help of a designed guide RNA, AMBER introduced on-target C-to-U editing on the eGFP transcript, and restored the correct codon and recovered eGFP fluorescence. These results confirmed that ADAR2 can be reprogrammed into a sequence-specific C-to-U RNA deaminase.
The team then evaluated AMBER across multiple endogenous targets and exogenous disease-related mutant targets in different cell lines. They systematically optimized guideRNA design, including length, the sequence context around the target cytidine, and strategies to reduce bystander editing in the editing window. Through this work, they established design principles that support high-efficiency and high-precision C-to-U editing while minimizing off-target events.
To assess AMBER’s therapeutic potential in vivo, the researchers delivered DNA constructs encoding AMBER into mice via tail-vein injection. Bioluminescence imaging of a Firefly luciferase (Fluc) reporter co-expressed with AMBER indicated that the editor accumulated mainly in the liver. RNA sequencing (RNA-seq) of liver tissue showed robust C-to-U editing at intended RNA targets. Meanwhile, transcriptome-wide analysis revealed minimal off-target editing events, and global gene expression was minimally affected when compared with control mice. These data indicate that AMBER can perform efficient and relatively specific RNA editing in vivo.
AMBER represents a new class of human-derived C-to-U RNA base editors with several advantageous properties: small molecular size, low immunogenicity, and high on-target editing efficiency. By avoiding bacterial CRISPR-Cas components and DNA/RNA dual-active APOBEC enzymes, AMBER successfully overcomes key barriers that previously limited in vivo C-to-U RNA base editing.
Compared with DNA editing approaches, RNA base editing has the capability to rewrite genetic information while offering improved safety due to its reversibility as well as its temporal and spatial tunability. This reversibility, combined with its safety profile in mouse liver, suggests that AMBER could be a practical platform for developing RNA editing therapies for genetic diseases caused by pathogenic T-to-C mutations. This work also expands the toolbox of RNA editors and offers a new technical route for translating RNA editing into clinical applications.
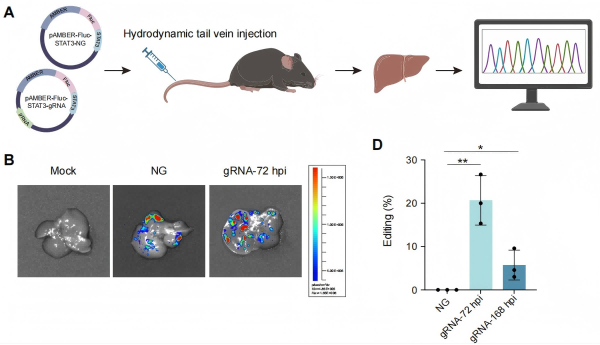
Figure: Expression and functional validation of the AMBER editor in mouse liver. (A) Schematic of in vivo delivery and editing validation of AMBER; (B) Verification of successful delivery and expression of the editing system in liver tissue using the Fluc signal co-expressed with AMBER; (C) Editing efficiency at the target site in liver tissue at 72 h and 168 h after injection. (Image by HAO Pei’s and LI Xuan’s groups)
Link: https://www.pnas.org/doi/10.1073/pnas.2505269122
Contact:
DIAO Wentong
Shanghai Institute of Materia Medica
E-mail: diaowentong@simm.ac.cn




